
吳亭穎 (Wu, Ting-Ying)
助研究員
- 瑞士蘇黎世聯邦理工學院生物學系 博士
- 基因和訊息傳遞網絡在植物對抗非生物逆境下的演化和反應機制,植物系統生物學,體學
- tingying@gate.sinica.edu.tw
- tingying@as.edu.tw
- +886-2-2787-1070 (Lab: R425)
- +886-2-2787-1090 (Office: R427)
- Lab Website
- ORCID
- Google Scholar
Our research: Plant-environment interaction in land plant evolution
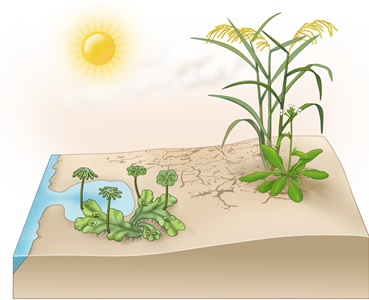
During plant terrestrializaiton (moving from water to land environments), plants face many terrestrial stressors, such as drought, high salinity and drastic temperature shifts. The main goal of our research is to understand how land plants adapt to those unfavoured environments at the molecular level during the course of evolution. We are also interested in applying the knowledge we obtained in such studies for designing stress resilient crops for precision agriculture.
Our approaches: Experimental systems biology
Multi-omics: By using bulk RNA-seq, single-cell RNA-seq, ATAC-seq and proteomics, we can accurately measure and quantify the genes and/ or proteins changes at a system level. By harnessing the hidden information from these big data, it also allows us to infer hypothesis and answer new questions.
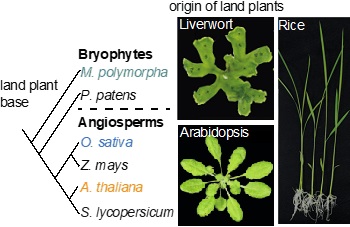
Simple phylogeny of land plants
Multi-species: By combining the experimental data from diverse model plant species such as Arabidopsis thaliana (dicot and vascular plant), Oryza sativa (monocot and vascular plant) and Marchantia polymorpha (bryophyte and non-vascular plant), we can study the molecular evolution of distinct or conserved gene regulatory network (GRNs) and signaling networks in the green lineage.
Multi-networks: Integrated network analysis is crucial for accurately predicting regulatory circuit. Gene regulatory networks (GRNs) allow us to understand the dynamics and hierarchy of hundreds and thousands of regulators (e.g., Transcription factors, TF) and their targeted genes simultaneously. Likewise, signaling networks will provide us a sophisticated understanding of kinase cascade dynamics upon environmental perturbations.
Current research questions:
- Understanding chromatin accessibility dynamics and gene regulatory landscape of multiple abiotic stress response in long-term plants evolution
- Understanding global phosphorylation landscape across distant species upon environmental perturbations
- Decoding plant abiotic response at the single cellular resolution in early land plant evolution
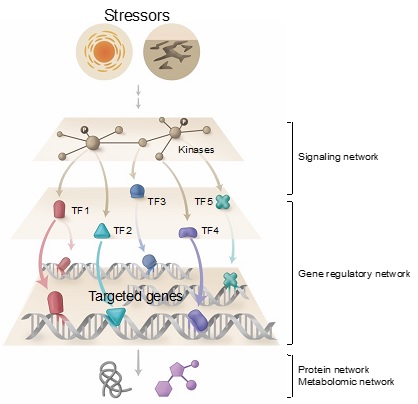
Different networks in plants
All publications
Selected Publications
- Wu TY*, Hor KL, Boonyaves K, Krishnamoorthi S and Urano D. Diversification of heat shock transcription factors expanded thermal stress responses during early plant evolution (2022). The Plant Cell, 00:1-20. *Corresponding author
- Boonyaves K, Wu TY, Dong YT and Urano D. Interplay between ARABIDOPSIS Gβ and WRKY transcription factors differentiates environmental stress responses (2022). Plant Physiology, 190(1):813-827.
- Akshaya K. Biswal, Ting-Ying Wu, Urano Daisuke, Pélissier Rémi, Jean-Benoit Morel, Alan M. Jones and Ajaya K. Biswal. Novel mutant alleles reveal a role of the extra-large G protein in rice grain filling, panicle architecture, plant growth, and disease resistance (2022). Frontier in Plant Science, 12:782960.
- Wu TY*, Goh HZ, Azodi C, Krishnamoorthi S, Liu MJ, and Urano D*. Evolutionarily conserved hierarchical gene regulatory networks for plant salt stress response (2021). Nature Plant, 7:787-799. *Co-corresponding author
- Wu TY, Mular M, Gruissem W, Bhullar NK. Genome Wide Analysis of the Transcriptional Profiles in Different Regions of the Developing Rice Grains (2020). Rice. 13, Article number: 62.
- Wu TY., Krishnamoorthi S., Richalynn L., Urano D. Transcriptional crosstalk between heterotrimeric G protein and nutrient stresses regulates plant adaptation to suboptimal micronutrient conditions (2020). Journal of experimental botany 71 (10), 3227-3239.
- Richalynn L., Wu TY., Jones A., Urano D., Quantitative morphological phenomics of rice and Arabidopsis G protein mutants are not consistent with a direct regulatory role in development. (2019), Developmental Biology, 457 (1), 83-90.
- Mangel N, Fudge JB, Li KT, Wu TY, Tohge T, Fernie AR, Szurek B, Fitzpatrick TB, Gruissem W, Vanderschuren H. Enhancement of vitamin B6 levels in rice expressing Arabidopsis vitamin B6 biosynthesis de novo genes (2019). Plant J., 99(6):1047-1065.
- Shih YJ, Chang HC, Tsai MC, Wu TY, Wu TC, Kao P, Kao WY and Chang IF (2018). Comparative leaf proteomic profiling of salt-treated natural variants of Imperata cylindrica. Taiwania, vol.63 no.2 pp.171-18.
- Wu TY, Gruissem W, and Navreet Bhullar. Targeting intracellular transport combined with efficient uptake and storage significantly increases grain iron and zinc levels in rice (2018). Plant Biotechnol J, https://doi.org/10.1111/pbi.12943
- Wu TY, Gruissem W, and Navreet Bhullar (2018). Facilitated citrate-dependent iron translocation increases rice endosperm iron and zinc concentrations. Plant Science, 270 (2018) 13–22.
- K. Boonyaves, Wu TY, Gruissem W, and N.K.Bhullar (2017). Enhanced grain iron levels in rice expressing an IRON-REGULATED METAL TRANSPORTER, NICOTIANAMINE SYNTHASE and FERRITIN gene cassette. Frontier in Plant Science. 8:130.
- Wu, T.Y., Kao, P., Chang, C.L., Hsu, P.H., Chou, C.H., Chang, I,.F. (2015). Phosphoproteomic profiling of microsomal fractions in leaves of Cogon grass (Imperata cylindrica). Plant OMICS. 8(6):595-603.
- Wu TY1, Juan YT1, Hsu YH, Wu SH, Liao HT, Fung RW, Charng YY (2013). Interplay between Heat Shock Proteins HSP101 and HSA32 Prolongs Heat Acclimation Memory Posttranscriptionally in Arabidopsis. Plant Physiol 161: 2075-2084. 1Co-first author.
Review article:
- Wu TY and Daisuke Urano. Genetic and systematic approaches toward G protein-coupled abiotic stress signaling in plants (2018). Frontier in Plant Science, 9.
Book Chapter:
- Ping Kao, Ting-Ying Wu and Ing-Feng Chang (2011). “Decreasing of Population Size of Imperata cylindrica Mangrove Ecotype and Sea-Level Rising.” Global Warming Impacts- Case Studies on the Economy, Human Health and on Urban and National Environments, Chapter 12, ISBN: 978-953-307-785-7.
國內
- 2024 前瞻計畫 - 中央研究院